Journal of Animal Research: v.10 n.1, p. 59-65. February 2020
DOI: 10.30954/2277-940X.01.2020.8
Detection and Identification of Mycobacterium tuberculosis and Mycobacterium bovis from Blood and Milk of Bovines
ABSTRACT
Bovine tuberculosis, a chronic disease of animals is caused by species of Mycobacterium tuberculosis complex (MTC) and it remains a potential threat to animals as well as humans. Differentiation of the species of MTC is required for epidemiological and diagnostic purpose. The present study evaluated the presence of different species of MTC in bovines using gyrB-restriction fragment length polymorphism analysis. In this study, blood and milk samples from 50 milch animals which were positive reactors of comparative intradermal tuberculin test were collected. Screening of MTC was done by IS6110-PCR using primers INS1/INS2 specific for MTC. The positive samples were further identified using gyrB- Restriction fragment length polymorphism analysis. Out of 50 positive reactors to CITT, only 4 (8%) animal were positive for MTC by IS6110-PCR. And gyrB-RFLP analysis using RsaI and SacII showed two positive for M. bovis and two animals for M. tuberculosis. Thus, gyrB-RFLP could be used as an additional tool in differential diagnosis of mycobacterial diseases thereby able to differentiate species of MTC.
Keywords: Mycobacterium bovis, Mycobacterium tuberculosis complex, PCR, RFLP.
Bovine tuberculosis, a chronic disease of animals is caused by species of Mycobacterium tuberculosis complex and it remains a potential threat to animals as well as humans. Bovine tuberculosis can affect practically all mammals, causing a general state of illness, coughing and eventual death (OIE, 2009)Mycobacterium tuberculosis complex comprises of the closely related species M. tuberculosis, M. bovis, M. africanum, M. microti, and M. canetti. These species are the causative agents of tuberculosis (TB) in humans and animals. MTC are genetically closely related though differ in terms of host predilection, pathogenicity, epidemiology and geographical distribution range (Chimara et al., 2004). The disease is distributed worldwide and in India, 300,000 people die from TB annually (TB India, 2012). The disease is of economic and public health importance. In developing countries, 10-15% of human TB while about 1-2% in developed countries is caused by M. bovis (Ashford et al., 2001).
Early diagnosis of mycobacterial infections plays a vital role in control of tuberculosis. Diagnosis of bovine tuberculosis is based primarily on conventional methods (culture, acid fast bacilli staining tuberculin test) and recently on molecular techniques (Filia et al., 2016; Langnyei et al., 2016). Though culture is the gold standard for diagnosis of tuberculosis, it takes several weeks to identify the species of Mycobacterium (Figueiredo et al., 2009). Tuberculin test is widely used for diagnosis of bovine tuberculosis since it is easy to perform on livestock in a large scale. But the disadvantage of this test is that it lacks both having specificity and sensitivity (Figueiredo et al., 2009).
How to cite this article: Gill, A.K., Filia, G., Leishangthem, G.D., Narang, D., Singh, S.T. and Singh, C.K. (2020). Detection and identification of Mycobacterium tuberculosis and Mycobacterium bovis from blood and milk of bovines. J. Anim. Res., 10(1): 59-65.
Various PCR-mediated methods have been developed for the rapid detection and differentiation of mycobacterial species. One of these methods is PCR amplification of a fragment of gyrB gene followed by restriction digest of the PCR products. PCR-RFLP of the gyrB gene using the commonly used enzyme Rsal permits one to easily separate the MTBC species from other atypical Mycobacterium.The present study was designed with the objectives to detect and differentiate members of Mycobacterium tuberculosis complex in bovines.
MATERIALS AND METHODS
Ethical approval
This study was approved by animal ethics committee of Guru Angad Dev Veterinary and Animal Sciences University (GADVASU) (No. IAEC/2016/ 643-675 dated 19/10/2016).
Screening of animals by Comparative Intradermal Tuberculin Test (CITT)
Screening of animal was done from five different organised and unorganised farms in Ludhiana, Kapurthala, and Moga districts of Punjab. A total of 190 animals (131 cattle and 59 buffaloes) more than three years of age from organized and unorganised dairy farms were screened for the cell mediated immune response by Comparative Intradermal Tuberculin test (CITT) (OIE 2009). CITT was performed as per guidelines of OIE (2009)using bovine tuberculin PPD from culture ofM. bovis (strain AN5, 3000 IU) and avian tuberculin PPD from culture of M. aviumsubspecies avium (strain D4ER, 2500 IU), obtained from Prionics (Switzerland). Bovine and avian tuberculins PPD (0.1ml each) were injected intradermally in the cervical area at least 12cm apart. The correction of the injection was confirmed by palpation of a small pea like swelling at the site. Inflammatory responses were recorded 72 hrs post injection. Animals were considered as positive reactors if the increase in skin thickness at the bovine site of injection was 4 mm or greater than the reaction at the site of the avian injection while no or ˂ 1 mm difference in the increase in skin fold reaction was considered negative reactors. Difference between 1-4 mm thicknesses was considered as inconclusive.
Extraction and detection of DNA from blood and milk samples
Whole blood samples (5ml) from all the 50 animals positive by CITT were subjected to DNA extraction using DN easy Blood and Tissue kit (Qiagen). One ml of blood was taken and centrifuged at 14,000 rpm for 15 min. The pellet was suspended in lysis buffer, and further DNA extraction was done as per the manufacturer’s protocol. DNA was eluted and stored at -20°C till further use.
Milk sample (20 ml) was transferred to centrifuge tube and centrifuged at 4000 rpm for one hour. After centrifugation whey was discarded while pellet (SNF) and cream were pooled and collected in 1.5 ml eppendorf tube. Pooled cream and SNF were transferred to column of 0.9 per cent HPC and were subjected to decontamination with constant shaking at 120 rpm in shaking incubator at room temperature for 4 hours. The tubes were kept undisturbed overnight at room temperature to allow the separation of cream and SNF. After separation, HPC was discarded and cream and SNF were pooled together in 1.5 ml eppendorf tube. Pooled mixture was given four washings with 1ml PBS by centrifugation at 5000 rpm, each washing was of 5 min. DNA was extracted from the mixture by using both conventional (Van Soolingen et al., 1997) and Kit method (PROMEGA, Wizard® Genomic DNA Purification Kit).
Polymerase chain reaction (PCR)
PCR reactions were performed on both blood and milk DNA (Hermans et al., 1990; Rodriguez et al.,1999). Reaction mixture (25 µl) containing 12.5 pl of Taq PCR master mix (Qiagen), 5 pl of DNA template and 0.2 µM of each primers INS1 (forward) 5’-CGTGAGGGCATCGAGGTGGC-3’, INS2 (reverse) 5’-GCGTAGGCGTCGGTGACAAA- 3’ for MTC. Along with sample DNA, a known positive control DNA and a negative control was also amplified. Thermal cycling was performed in T Gradient Thermocycler (Biometra, Germany) with the following cycling parameter for INS1 and INS2: 94 for 5 min followed by 30 cycles of 1 min at 94°C, 1 min at 63°C and 1 min at 72°C followed by final extension of 7 min at 72°C. PCR products were run by agarose gel electrophoresis using 1.5% agarose gel and visualized by gel documentation system (Bio-Rad).
Amplification by gyrB-PCR
The target DNA for amplification was 1,020 bp fragment of the gyrB gene, which was used to identify members of the M. tuberculosis complex. The primers used were MTUB-f (5’-TCG GAC GCG TAT GCG ATA TC-3’) and MTUB-r (5’-ACA TAC AGT TCG GAC TTG CG- 3’) (Chimara et al., 2004; Niemann et al., 2000) which amplify a gene sequence gyrB for 1,020 base pair fragment in MTC PCR. Briefly, a ready to use TaqPCR Master Mix (Qiagen) was used. A reaction volume of 50 µl was made, containing 25 pl of TaqPCR Master Mix, 2 pl of forward primer (10 pmol/pl), 2µl of reverse primer (10 pmol/pl), 11pl of nuclease free water and 10 pl of DNA template. Thermal cycling was performed in T. Gradient Thermo cycler (Biometra, Germany). The cycling conditions were as follows, initial denaturation at 95 °C for 10 min, followed by 35 cycles for denaturation at 94 °C for 1 min, annealing of primers at 65 C for 1 min, extension at 72 C for 1.5 min and final extension at 72 C for 7 min. PCR products were run by gel electrophoresis and visualized by gel documentation system (Bio-Rad).
Restriction fragment length polymorphism (RFLP)
DNA polymorphisms in the 1,020-bp gyrB fragment amplified with the primers MTUB-f and MTUB-r were analyzed after digestion with restriction enzymes RsaIand SacII as recommended by the manufacturer’s protocol (Promega).Digestion mixture consist of 16.3 pl sterile, deionized water, 2 pl RE 10X buffer, 0.2 pl of 10 pg/ pl Acetylated BSA and 1 pl of 1 pg/pl DNA, mixed by pipetting followed by 0.5 pl of 10 pg/pl Restriction Enzyme. The mixture was incubated at 37 °C for 4 hours. The DNA digest was separated in a 2% agarose gel by electrophoresis, and the gels were visualized under a UV transilluminator. A 100-bp DNA ladder was used as a size standard
RESULTS AND DISCUSSION
Mycobacterial infection is considered as a potential threat to both animal and human health. Detection and identification of mycobacterial species were done basically by conventional AFB staining and culture which are usually time consuming and also unable to differentiate M. tuberculosis from other non- tuberculosis Mycobacterium. PCR techniques offer high sensitivity and have been successfully used for diagnosing bovine TB in several types of naturally infected organic materials such as tissue, blood, and nasal exudates (Filia et al.,2016; Langnyei et al., 2016; Coetsie et al., 2000; Ahir et al., 2016).
Diagnosis of bovine tuberculosis in live animals is primarily based on the detection of specific cell-mediated immune (CMI) responses and molecular techniques. In recent years, various attempts have been made to develop alternative and more rapid methods of detection of MTC. In this study, out of 190 animals tested by CITT, 70 (36.84%) animals were positive, 41 (21.57%) inconclusive and 79 (41.57%) were negative reactors. From these 70 positive animals, blood and milk samples from 50 milch animals were screened for Mycobacterium tuberculosis complex using INS1/INS2 primers followed by PCR-RFLP. Out of these 50 blood samples, 4 (8%) were positive for MTC (Figure 1) while no milk samples were positive for MTC. The results in the present study are similar to those reported by other workers in organized farms using single intradermal tuberculin test and CITT (Filia et al., 2016; Ahir et al., 2016; Thakur et al., 2010).
For the rapid and specific diagnosis of TB, PCR assays are the most promising alternative method (Figueiredo et al., 2009; Serrano-Moreno et al., 2008). PCR techniques offer high sensitivity and have been successfully used for diagnosing bovine TB in several types of naturally infected organic materials such as tissue, blood, and nasal exudates (Coetsie et al., 2000; Gomez-Laguna et al., 2010). The most commonly used PCR is based on primers that amplify segments of the IS6110 element, particularly targeting 245 bps fragments. In this study, blood and milk samples from 50 milch animals were screened for Mycobacterium tuberculosis complex by IS6110 PCR amplify an insertion sequence of 245 bps using INS1/INS2 primers followed by PCR-RFLP. Out of these 50 blood samples, 4 (8%) were positive for MTC while no milk samples were positive for MTC.The results in the present study are similar to those of Filia et al. (2016) who observed that 4.23% blood samples were positive by PCR. Basit et al. (2015) reported the overall prevalence of tuberculosis was 6.5% by PCR from a total 200 tissue samples of lungs, lymph nodes and liver from cattle, buffaloes, sheep and goats collected from abattoir.
In the present study, as compared to CITT, only 4 blood samples were positive by PCR. CITT has been considered less in sensitivity and specificity. Though PCR-based methods helps in rapid detection of target DNA in suspected clinical samples, the efficacy of PCR assays can be influenced by the quality of target DNA and the presence of sample-derived PCR inhibitors (Schrader et al., 2012). PCR inhibitors found in blood, serum or plasma samples includes substances like IgG, haemoglobin and lactoferrin (Al-Soud et al., 2000; Al-Soud et al., 2001). Most of these inhibitors are presumed to affect the RNA directly and not the enzymes of the reaction (Konet et al.,2000).
However, in the present study, milk samples were found negative by IS6110 PCR. The absence of Mycobacteriumspecies in the milk samples from CIIT-reactive cows by PCR might be due to onetime sampling in the design of the study where some periods of bacterial excretion might have been missed (Carvalho et al., 2014) or due to presence of few numbers of excreted bacteria, or to the presence of dead or non-viable bacilli due to the action of macrophages, or even to the use of the decontamination method (Zumarraga et al., 2006). The intermittent character of bacilli secretion after a short constant post¬infection period was also documented (Menzies et al.,2000).
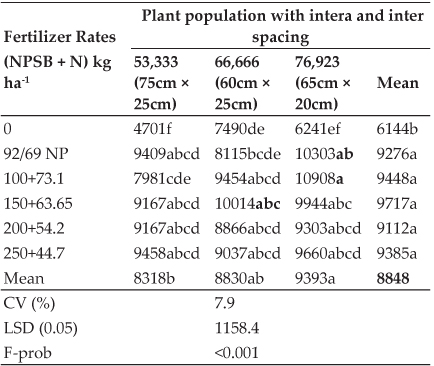
For further confirmation of the species of Mycobacterium tuberculosis complex, gyrB PCR restriction endonuclease differentiation was carried out. For gyrB-RFLP, these 4 positive DNA samples were further amplified by PCR using the primer pair MTUB-f and MTUB-r specific for amplification of the 1,020-bp fragment of the gyrB gene of the M. tuberculosis complex (Fig. 2). These four PCR products along with a positive control were digested for 4 hours at 37°C with restriction enzyme RsaI and SacII. These enzymes digest the DNA of specific species and cut at the restriction site specific for these enzymes. GyrB- RFLP revealed RsaI digestion showing bands at 480bps and 385bps forM. bovis and at 560bps and 385bps forM. tuberculosis. SacII shows no digestion confirming presence of M. bovis not M. caprae. Agarose gel electrophoresis of RFLP is shown in Fig. 3.

Fig. 1: Agarose gel electrophoresis of the amplified IS6110 gene using INS1/INS2 primers
M: 100 bp plus DNA ladder; L1: Positive control; L2: Negative control; L3-L6: samples
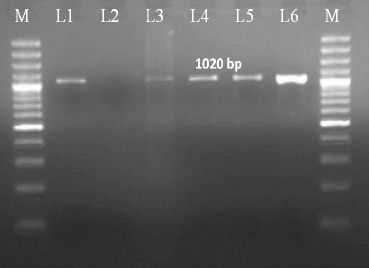
Fig. 2: Agarose gel electrophoresis of the amplified GyrB gene by using MTUB1/MTUB2 primers
M: 100 bp plus DNA ladder; L1: Positive control; L2: Negative control; L3-L6: samples
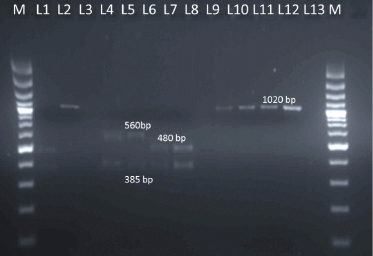
Fig. 3 : RFLP patterns of PCR products obtained by Rsal digestion and SacII digestion
M: 100 bp plus DNA ladder; L1: Positive control digested by RsaI; L2: Positive control digested by SacII; L4-L7: Samples digested by RsaI; L9-L12: Samples digested by SacII
PCR-RFLP for detection and differentiation of tuberculosis with selective amplification of the gyrB fragment from each species of the M. tuberculosis complex were done by various workers (Chimara et al., 2004; Niemann et al, 2000; Abass et al, 2010; Kasai et al, 2000; Goh et al., 2006). In the present study, gyrB-RFLP showed that by RsaI digestion, bands at 480 and 385 base pair were observed in two samples indicating M. bovis while 560 and 385 in another two samples indicating M. tuberculosis.Further SacII enzyme digestion was done to differentiate M. bovis subspecies i.e., M. bovis subsp. bovis and M. bovis subsp. caprae. No digestion was observed in all the samples and this lack of digestion with SacII excluded the presence of M. bovis subsp. caprae confirming the presence ofM. bovis subsp. bovis.
For differentiation of strains of M. tuberculosis, M. bovis, M. africanum and M. microti Kasai et al. (2000) developed gyrB-RFLP. It was found that 1020 bp fragment of the gyrB gene was amplified with specific primers, which do not generate amplicons from other species of mycobacteria followed by digestion of the amplicon with restriction enzymes. Chimara et al. (2004) analyzed gyrB- RFLP patterns from reference strains which include M. tuberculosis H37Rv, M. bovis, M. bovis BCG, M. canetti, M. africanum and M. pinnipedii isolates. They developed a new table based on band sizes and confirmed by analysis of sequences deposited by Kasaiet al. (2000). The band pattern of the present gyrB-RFLP was similar to that observed by Chimara and his groups (2004) but differ from Niemann et al. (2000) where bands showed different sizes. In our study small band as described by Chimara et al. (2004) were not observed. These small bands were also not considered by other workers (Niemann et al., 2000; Kasai et al., 2000).
In the present study, M. tuberculosis and M. bovis were detected in animals located at different areas. Moreover, two animals were positive for M. tuberculosis indicating that the infection was caused from human contact which may be animal handlers indicating a reverse zoonosis in this areas. Many workers have reported that animals can be infected by exposure to any individual shedding the organism (Ayele et al., 2004; Fritsche et al., 2004; Grange et al., 1994). Animals infected with M. tuberculosisconstitute a potential hazard of transmission of virulent tubercle bacilli back to humans (Mishra et al., 1997). The presence of MTC in animal may raise concerns regarding the zoonotic risk for humans, especially those living at the animal-human interface.
CONCLUSION
Thus, GyrB-RFLP showed promising results in diagnosis and further differentiation of MTC in early stage in live animals. GyrB-RFLP serves as an additional tool for differentiation of MTC and may help in the epidemiological studies. This can also help in quick segregation of infected animals; restrict transmission and rapid eradication of bovine TB in the country. Further large-scale studies are required for detection, control and eradication of bovine TB in the state.
Acknowledgements
The authors acknowledged Director Research, GADVASU for providing funds through Rashtriya Krishi Vikas Yojna for the study.
Conflict of Interest: The authors declared no conflict of interest.
REFERENCES
OIE. 2009. Bovine Tuberculosis. OIE Terrestrial Manual.Paris1-16.
Chimara, E., Ferrazoli, L. and Leao, S.C. 2004. Mycobacterium tuberculosis complex differentiation using gyrB restriction fragment length polymorphism analysis. Mem. Inst. Oswaldo Cruz. Rio de Janeiro., 99 : 745-748.
TB India.2012. Revised National TB control programme. Annual status report, New Delhi India, Available from, http// www.tbcindia.nic.in/ [Accessed on 20-07-2015].
Ashford, D.A., Whitney, E., Raghunathan, P. and Cosivi, O. 2001. Epidemiology of selected mycobacteria that infect humans and other animals. Rev. Sci. Tech., 20 : 325-337.
Filia G., Leishangthem G.D., Mahajan V. and Singh A. 2016. Detection ofMycobacterium tuberculosis and Mycobacterium bovis in Sahiwal cattle from an organized farm using ante¬mortem techniques. Vet. World, 9 : 383-387.
Langnyei, L.P., Leishangthem, G.D., Narang, D., Filia, G., Gupta, K. and Singh, A. 2016. Molecular and immunohistochemical detection of Mycobacterium bovis in formalin-fixed tissues from animals with spontaneous bovine tuberculosis. Indian J. Vet. Pathol., 40 : 116-121.
Figueiredo, E.E.S., Silvestre, F.G., Campos, W.N., Furlanetto, L. V., Medeiros, L., Lilenbaum, W., Fonsecal, L.S., Silva, J.T. and Paschoalin, VM.F. 2009. Identification ofMycobacterium bovis isolates by a multiplex PCR. Braz. J. Microbiol., 40 : 231-233.
Van Soolingen, D., Hoogenboezem, T., Haas, P.E., Hermans, P.W., Koedam, M.A., Teppema, K.S., Brennan, P.J., Besra, G.S., Portaels, F., Top, J., Schouls, L.M. and Van Embden, J.D.A. 1997. Novel pathogenic taxon of the Mycobacterium tuberculosis complex, Canetti, characterization of an exceptional isolate from Africa. Int. J. Syst. Bacteriol., 47 : 1236-1245.
Hermans, P.W., Soolingen, D.V., Dale, J.W., Schuitema, A.R.J., Mcadam, R.A., Catty, D. and Vanembden, J.A. 1990. Insertion element IS986 from Mycobacterium tuberculosis, a useful tool for diagnosis and epidemiology of tuberculosis. J. Clin. Microbiol., 28 : 2051-2058.
Rodriguez, G.M., Gold, B., Gomez, M., Dussurget, O. and Smith, I. 1999. Identification and characterization of two divergently transcribed iron regulated genes in Mycobacterium tuberculosis. Tuber. Lung Dis., 79 : 287-298.
Niemann, S., Harmsen, D., Sch-gerdes, S.R. and Richter E. 2000. Differentiation of clinical Mycobacterium tuberculosiscomplex isolates by gyrB DNA sequence polymorphism analysis. J Clin. Microbiol., 38 : 3231-3234.
Abass, N.A., Suleiman, K.M. and Jalii, I.M.E.I. 2010. Differentiation of clinical Mycobacterium tuberculosiscomplex isolates by their gyrB polymorphism. Indian J. Med. Microbiol., 28 :26-29.
Kasai H., Ezaki, T. and Harayama, S. 2000. Differentiation of phylogenetically related slowly growing mycobacteria by their gyrB Sequences. J. Clin. Microbiol., 38 : 301-308.
Coetsier, C., Vannuffel, P, Blondeel, N., Denef, J F., Cocito, C.and Gala, J. L. 2000. Duplex PCR for differential identification of Mycobacterium bovis, M. avium, and M. avium subsp. paratuberculosis in formalin-fixed paraffin-embedded tissues from cattle. J. Clin. Microbiol., 38 : 3048-3054.
Ahir, P, Filia, G., Mahajan, V., Leishangthem, G.D., Rai, T.S. and Singh, A. 2016. Diagnosis of Bovine tuberculosis in lactating cattle and buffaloes by comparative intradermal tuberculin test and bovine gamma-interferon. J. Anim. Res., 6: 1069-1072.
Thakur, A., Sharma, M., Katoch, V.C., Dhar, P. and Katoch, R. C. 2010. A study on the prevalence of bovine tuberculosis in farmed dairy cattle in Himachal Pradesh. Vet. World, 3 : 409-414.
Serrano-Moreno, B.A., Romero, T. A., Arriaga, C., Torres, RA., Pereira-Suarez, A.L.and Garcia-Salazar, J.A. 2008. High Frequency of Mycobacterium bovis DNA in colostra from tuberculous cattle detected by Nested PCR. Zoonoses Public Health, 55 : 258-266.
Gomez-Laguna, J., Carrasco, L., Ramis, G., Quereda, J.J., Gomez,S. and Pallares F.J. 2010. Use of real-time and classic polymerase chain reaction assays for the diagnosis of porcine tuberculosis in formalin-fixed, paraffin-embedded tissues. J. Vet. Diagn. Investig., 22 : 123-127.
Basit, A., Hussain, M., Ayaz, S., Shahid, M., Rahim, K., Ahmad, I., Ullah, R., Hashem, A., Abd-Allah E., Alqarawi A. A.and Gul N. 2015. Isolation and identification of Mycobacterium bovis and Mycobacterium tuberculosis from animal tissues by conventional and molecular method. Indian J. Anim. Res.,49: 687-693.
Schrader, C., Schielke, A., Ellerbroek, L. and Johne R. 2012. PCR inhibitors - occurrence, properties and removal. J. Appl. Microbiol, 113 : 1014-1026.
Al-Soud, W.A.and Radstrom P. 2000. Effects of amplification facilitators on diagnostic PCR in the presence of blood, feces, and meat. J. Clin. Microbiol., 38 : 4463-4470.
Al-Soud, W.A. and Radstrom, P. 2001. Purification and characterization of PCR-inhibitory components in blood cells. J. Clin. Microbiol., 39 : 485-493.
Konet, D.S., Mezencio, J.M., Babcock, G.and Brown, F. 2000. Inhibitors of RT-PCR in serum . J. Virol. Methods, 84 : 95-98.
Carvalho, R.C.T., Castro, V.S., Fernandes, D.V.G.S., Moura, G., Celestina, F. Santos, E.C. Paschoalin V.M.F. and Figueiredo, E.E.D.S. 2014. Use of the PCR for Detection Mycobacterium bovis in Milk . Blucher Food Sci. Proceedings, 1 : 359-360.
Zumarraga, M.J., Meickle, V., Bernardelli, A., Abdala, A., Tarabla, H., Romano, M.I. and Cataldi, A. 2006. Use of touch¬down polymerase chain reaction to enhance the sensitivity of Mycobacterium bovis detection. J. Vet. Diagn. Investig., 17 : 232-238.
Menzies, F.D. and Neill, S.D. 2000. Cattle-to-cattle transmission of bovine tuberculosis. Vet. J., 160 : 92-106.
Goh, K.S., Fabre, M., Huard, R.C., Schmid, S. and Rastogi, S.C. 2006. Study of the gyrB gene polymorphism as a tool to differentiate among Mycobacterium tuberculosis complex subspecies further underlines the older evolutionary age of Mycobacterium canettii. Mol. Cell Probes., 20 : 182-190.
Ayele, W.Y., Neil, S.D., Zinsstag, J, Weiss, M.G. and Pavlik, I. 2004. Bovine tuberculosis, an old disease but a new threat to Africa. Int. J. Tuberc. Lung Dis., 8 : 924-927.
Fritsche, A., Engel, R., Buhl, D. and Zellweger, J.P. 2004. Mycobacterium bovis tuberculosis, from animal to man and back. Int. J. Tuberc. Lung Dis., 8 : 903-904.
Grange, J.M. and Yates, M.D. 1994. Zoonotic aspects of Mycobacterium bovis infection. Vet. Microbiol., 40 :137-151
.Mishra, J.K, Panda, S.N. and Panda, H.K. 1997. Prevalence of bovine tuberculosis among farm cattle in Orissa. Indian Vet. J, 74: 195-198.